Скачать с ютуб Using the ggplot theme function to customize facet labels and your legend (CC067) в хорошем качестве
Скачать бесплатно Using the ggplot theme function to customize facet labels and your legend (CC067) в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно Using the ggplot theme function to customize facet labels and your legend (CC067) или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон Using the ggplot theme function to customize facet labels and your legend (CC067) в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
Using the ggplot theme function to customize facet labels and your legend (CC067)
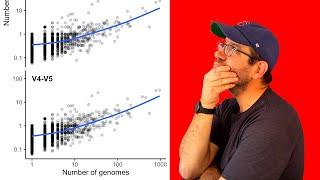
Have you ever wondered hot to use ggplot's theme function? In today's episode of Code Club, Pat Schloss shows you how you can use the theme function to customize figures you make with ggplot2. He'll show you how to use the theme function to modify the position and text in your facet labels. If that isn't enough, he then demonstrates how you can also use the theme function to move the position and sizing of your legend. This episode is part of a larger arc of episodes investigating the sensitivity and specificity of amplicon sequence variants (ASVs), also known as exact sequence variants (ESVs) and operational taxonomic units (OTUs). ASVs are growing in popularity for analyzing microbial communities using 16S rRNA gene sequences. Proponents think that they should supplant operational taxonomic units (OTUs). What do you think? Pat demonstrates these concepts by live coding at the command line interface using GitHub Flow, Make, and RStudio. The accompanying blog post can be found at http://www.riffomonas.org/code_club/2... You can also find complete tutorials for learning R with the tidyverse using... Microbial ecology data: http://www.riffomonas.org/minimalR/ General data: http://www.riffomonas.org/generalR/ 0:00 Introduction 1:50 Critique of lumping and splitting figure 3:45 First iteration of figure 11:13 Using facet labels as axis labels 19:08 Moving position of legend 20:40 Adjust space between legend keys 22:29 Inserting figure into manuscript 26:29 Conclusion