Скачать с ютуб Phage display - theory, techniques, & uses в хорошем качестве
Скачать бесплатно Phage display - theory, techniques, & uses в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно Phage display - theory, techniques, & uses или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон Phage display - theory, techniques, & uses в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
Phage display - theory, techniques, & uses
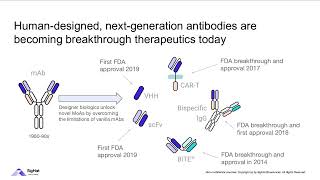
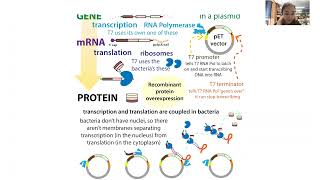
“Phage display” might sound a bit like some fancy new window display retailers put out to entice shoppers. It’s definitely not that - but in a way it is kinda like molecular “window shopping…” Phage display is a technique in which bacteria-infecting viruses called bacteriophages (“phages”) are used to “display” bits of proteins on their surface. You can get lots of phages to display lots of different protein bits and then add this “phage display library” to some molecule you want to see if those protein bits bind to. You then select for the best binders and figure out what they’re showing. full text & figures: blog form: http://bit.ly/phagedisplay Pharmaceutical companies and researchers often turn to phage display if they want to develop lab-made antibodies or peptide-based drugs. In fact, one of the world’s biggest-selling drugs, Humira (adalimumab) was the first FDA-approved human monoclonal antibody therapy and it was developed using phage display. More about that and other specific applications later, but first let’s talk about how it works. Basically, “genes” are stretches of DNA with instructions for making functional products like proteins (which are made up of protein “letters” or “building blocks” called amino acids connected through peptide bonds, hence the name “peptides” for protein bits). So, for example, phages have genes with DNA instructions for making “coat proteins” to form a protective coat around them when they travel and infect new cells. If you you stick foreign DNA into the genes of the phage coat proteins, an extra bit of protein (the amino acids corresponding to the inserted DNA) will get added into the phage coat protein. Kinda like if you have a recipe for a 3-layer cake and you sneak in an extra page with instructions for making a fourth layer. Since the coat protein sticks out from the surface of the phage, if you position the insert right you can get the extra bit “displayed” (sticking out to the environment). And then you can check to see if that displayed bit can bind something you’re interested in. The foreign DNA you insert can either be DNA for something you know you’re interested in, like an antibody fragment, or random DNA if you want to unbiasedly search for peptides. A common application, sometimes referred to as “biopanning,” is to coat wells of a plate with some binding partner, like maybe a protein you want to find an inhibitor for). And then you take a phage library and add it to the well. If the phage is displaying something that can bind that protein, that phage will stick. If not, the phage won’t, so you can wash the “losers” off. And then you can get the stuck phages to unstick and sequence them to see what DNA insert they have. Since you only have a few copies at that point, usually you first do an “amplification” step, where you let the phage infect some more bacteria and make more copies of themselves first before you try to go sequencing them. But you usually don’t even do the sequencing until you’ve gone through several cycles of “affinity selection,” where you make the wash conditions harsher and harsher each cycle. So the basic rundown is: - make phage library - stick foreign bits of DNA into phage coat proteins - these bits can be totally random or a “curated collection” such as instructions for pieces of antibodies - let those phages infect bacteria - the phages will grow and insert the modified proteins into their coats - add the phages to the sticking test (this can be wells of a plate, beads, even whole cells) - give them some time to stick - wash off the ones that didn’t stick (such as by increasing the salt concentration) - wash off the ones that did stick (such as by altering the pH) - amplify the ones that stuck - you start with a TON of phage, and only a small portion of them will bind (especially in the first round) so you need to make more if you want to test them more (and find out what they are) - let them infect more bacteria so they can make more copies of themselves - put those to the sticking test again, but this time use harsher wash conditions to keep only the stronger stickers - amplify them - do this again as many times as you want in order to find the strongest stickers - note: there will be some mutations naturally randomly introduced into the inserted DNA, so the bits might even get better than the bits you started with! (and you can promote this mutation by growing the phages in “mutator strains” of bacteria) - once you’re satisfied, sequence the phage coat protein insert - now that you know what it is, you can do different things with it. So, for example, if it was an antibody part you can insert that DNA into the rest of the antibody gene and get cells to make it for you. If it was a random peptide you found bound to a protein you want to drug, you can synthesize that peptide and then test to see if it inhibits the protein you found it bound.