Скачать с ютуб A little wobble hypothesis goes a long way: explaining codon-anticodon interactions в хорошем качестве
Скачать бесплатно A little wobble hypothesis goes a long way: explaining codon-anticodon interactions в качестве 4к (2к / 1080p)
У нас вы можете посмотреть бесплатно A little wobble hypothesis goes a long way: explaining codon-anticodon interactions или скачать в максимальном доступном качестве, которое было загружено на ютуб. Для скачивания выберите вариант из формы ниже:
Загрузить музыку / рингтон A little wobble hypothesis goes a long way: explaining codon-anticodon interactions в формате MP3:
Если кнопки скачивания не
загрузились
НАЖМИТЕ ЗДЕСЬ или обновите страницу
Если возникают проблемы со скачиванием, пожалуйста напишите в поддержку по адресу внизу
страницы.
Спасибо за использование сервиса savevideohd.ru
A little wobble hypothesis goes a long way: explaining codon-anticodon interactions
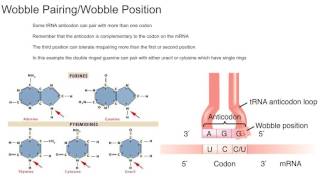
What is wobble? How do codons and anticodons really interact? What are the special rules that define the tRNAs used during translation? Find the answers in this review of: Codon--anticodon pairing: the wobble hypothesis. Crick FH. J Mol Biol. 1966 Aug;19(2):548-55. doi: 10.1016/s0022-2836(66)80022-0. Twitter: / genetics_stuff OTHER VIDEOS YOU MIGHT LIKE: • Industrial melanism: a black and white example of natural selection? (Kettlewell, 1955) - • Industrial melanism: a black and whit... • Classical vs Balance model of genetic variability: BALANCE FOR THE WIN (Lewontin & Hubby, 1966) - • Classical vs Balance model of genetic... • Transcription factor AGAMOUS: the molecular genetics of flower development (Yanofsky et al., 1990) - • Transcription factor AGAMOUS: the mol... In 1961, Marshall Nirenberg demonstrated that RNA was required to polymerise amino acids into protein. A year later, he showed synthetic RNA homopolymers comprising only of uracil enabled in vitro production of polypeptides containing phenylalanine, and those comprising only of cytosine allowed polypeptides containing proline to be synthesised. Nirenberg, Khorana and others subsequently tested all 64 possible codons for the amino acids they encoded, earning the 1968 Nobel Prize for Physiology and Medicine for the interpretation of the genetic code and its function in protein synthesis. Out of the 64 possible codons, only 61 encoded one of the twenty amino acids. Groups of two, three or four codons often encode the same amino acid, rending much of the genetic code redundant. This redundancy was not random either as synonymous codons only varied in their last nucleotide and followed three main patterns of redundancy. Those ending in C were generally synonymous with those in U, those in A were synonymous with G, or all four were synonymous with each other. Furthermore, Robert Holley and others demonstrated synonymous codons often bound the same tRNA molecule, implicating base pairing between tRNA and codon nucleotides in the 3rd position did not follow normal pairing rules. Having devised the original rules for base pairing, molecular biologist Francis Crick took great interest in understanding this unconventional interaction, proposing the wobble hypothesis as a possible explanation. Crick suggested that the first tRNA anticodon base, which interacts with the third codon base, ‘wobbled’ in three dimensional space, allowing it to form unconventional pairs alongside those permitted by standard Watson and Crick base paring. In normal pairing only two combinations are possible: between A and U, and C and G. Additional base pairings which the third nucleotide might observe are those between G and A, C and U, U and U, G and U, I and U, or I and A. Crick disqualified G and A, as it is unable to form sufficient hydrogen bonding, and both U and C, and U and U due to hindrance from glycosidic bonds and keto groups. Contrastingly, he deemed G and U, I and U, and I and A pairings possible as they could form necessary hydrogen bonding and didn’t require wobbling far from the standard position. Crick then evaluated possible codon-anticodon positions needed to enable these interactions, finding seven in total, although noting all seven are unlikely considering geometrical constraints during transcription. He disqualified any pairings which went against general features of the code, such as enabling multiple amino acids to be encoded by one codon. This left four total positions, including those of standard base-pairing, and so six in total. With wobble pairing, the minimum number of distinct tRNA anticodons needed by an organism is 32. In reality, the lowest number observed so far is 36 in the thermophilic archaea Methanocaldococcus jannaschii. Comparatively flies have 44 and humans 51. This variation in tRNA numbers is in part due to preferential use of particular synonymous codons over others. This “codon bias” varies by organism. In less complex organisms, areas of high G+C content favour synonymous codons ending in G+C. In more complex organisms, selective pressure for increased translation efficiency and accuracy is believed to favour some codons over others, but much is still unknown. Creator: Tobias Massang References: Nirenberg, M. (2004). Historical review: Deciphering the genetic code – a personal account. Trends Biochem Sci, 46-54. Iben, J. R., & Maraia, R. J. (2014). tRNA gene copy number variation in humans. Gene, 376–384. Crick, F. (1966). Codon—anticodon pairing: The wobble hypothesis. J Mol Biol, 548-555. International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature, 860–921. Rogers, H. H., Bergman, C. M., & Griffith-Jones, S. (2010). The Evolution of tRNA Genes in Drosophila. Genome Biol Evol, 467–477. Rocha, E. P. (2004). Codon usage bias from tRNA's point of view: Redundancy, specialization, and efficient decoding for translation optimization. Genome Res, 2279–2286.